Progress Report
Tuesday, November 30th, 2010
Progress and Agenda - Overview of Possibilities
- Triple-sandwich
- interesting possibility
- no longer on the table due to absence at the hospital
- Tagging
- awaiting more data from Leon
- synthetic data
- tracking method
- assessment method
- Analysis as above without tagging
Experiments - Recapitulation
- We need to obtain data, preferably with tags
- We need to synthesise data for validation
- Method of tracking
- Method of assessing cross-frame quality of fit
Framework

In KDE (Qt), despite MATLAB putting a lot of effort into improving the GUI on the GNU/Linux side, QtOctave integrates more nicely with the desktop. Some other tools of use include R, Sage, Scilab, QtOctave, and graphviz.
Framework - Ctd.
I test to see that code always works in both Octave and MATLAB. The syntax is compatible.

Framework - Ctd.
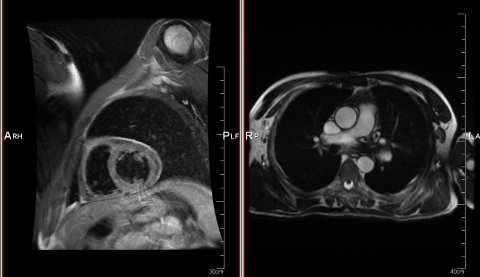
I'm also exploring the GPLv3-licensed BioSig project, which allows me to deal with DICOM datasets of the heart.
Framework - Ctd.
DICOM is considered "extensible", which implies that the usual type of format incompatibility will always occur. It's just an inconsistent data format.
Here is the command-line converter that I use (some are for Windows only) and some other options can be found on the Web through directories. There are also good software resources on scientific data formats and there is radiology CEU information regarding PACS and DICOM viewers.
Tagged Data
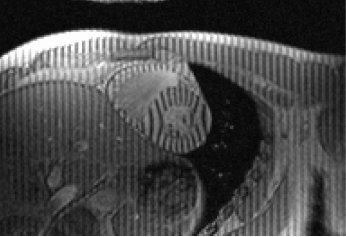
MRI tagging commonly used, not simple to find on the Web
Tagged Data - Ctd.
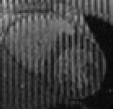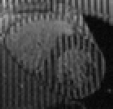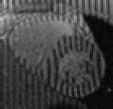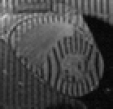
Based on the excellent Web page of Zhen Qian I have managed to piece together something for people to play with. From his thesis [PDF] I've extracted a series of 6 frames that I've produced the above videos from.
Tagging
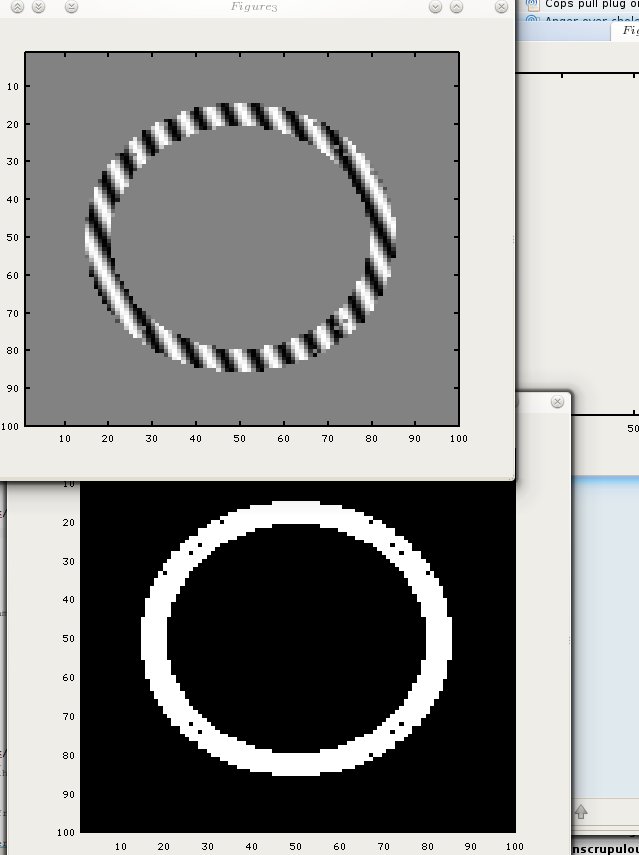
Tagging in the context of image acquisition involves getting magnetic signal by selectively sending charges to the tissue, whose atoms (usually just hydrogen is targeted by adjustment of frequencies) equipment can try to test for response in such a way that under normal conditions without noticeable change they would return a rectangular grid overlaid on top of the data. This enables better tracking of tissue motion that's robust to spatially-similar atoms (whose returned signal is hard to discern visually).
Future Directions
- Identify data worth working with
- synthetic
- NYU set
- existing sets of frames
- Methods to be determined
- fast implementation, selection of landmark points
- validation framework to measure performance