Wednesday, November 17th, 2010, 11:40 am
Gentle Background and Introduction to MRI Tagging

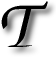 AGGING in the context of image acquisition involves getting magnetic signal by selectively sending charges to the tissue, whose atoms (usually just hydrogen is targeted by adjustment of frequencies) equipment can try to test for response in such a way that under normal conditions without noticeable change they would return a rectangular grid overlaid on top of the data. This enables better tracking of tissue motion that’s robust to spatially-similar atoms (whose returned signal is hard to discern visually). This post is a personal and informal quick survey of some resources which people may hopefully find helpful. It is not a detailed or comprehensive analysis.
AGGING in the context of image acquisition involves getting magnetic signal by selectively sending charges to the tissue, whose atoms (usually just hydrogen is targeted by adjustment of frequencies) equipment can try to test for response in such a way that under normal conditions without noticeable change they would return a rectangular grid overlaid on top of the data. This enables better tracking of tissue motion that’s robust to spatially-similar atoms (whose returned signal is hard to discern visually). This post is a personal and informal quick survey of some resources which people may hopefully find helpful. It is not a detailed or comprehensive analysis.
Background
Tagging has many areas about or around it which ought to be explored. Back in 2003, an extension of the approach — one of complementary innovation in particular — was a goal yet to be accomplished although many extensions already exist. As Professor Leon Axel from the Department of Radiology at NYU School of Medicine (he is the inventor of tagging) put it in his paper “Tagged MRI-Based Studies of Cardiac Function” (2003), improvements in tagged MRI methods include optimisation in terms of speed and 3-D as well. To quote, “[s]ome ways in which we are seeking to improve tagged imaging include improved physiologic motion synchronization, faster imaging methods and 3D image acquisition.”
In case magnetic resonance cannot be properly applied for 3-D tagged images, other modalities can perhaps be considered at an early stage where data to work on is being sought and gradually collected starting with coarse image sequences (more on that later). However, it is tagging which probably yields the most valuable information, adding it to one of the most valuable imaging modalities. As Wikipedia explains it, Speckle Tracking Echocardiography, for example, “analyzes motion within an ultrasonic window by tracking intereference patterns and natural acoustic reflections.[1] These reflections, also described as ‘‘speckles’’, ‘‘markers’’, ‘‘patterns’’, ‘‘features’’, or ‘‘fingerprints’’ are tracked consecutively frame to frame and ultimately resolved into angle-independent two-dimensional (2D) and three-dimensional strain-based sequences (3D) [2][3][4] These sequences provide both quantitative and qualitative information regarding tissue deformation and motion.”
The connection to MRI is mentioned further down where it says: “The utilities of STE are increasingly recognized. Strain results derived from STE have been validated using sonomicrometry and tagged MRI and results correlate significantly with tissue Doppler–derived measurements.[7][8][9]”
Reference 8 is “Amundsen BH, Helle-Valle T, Edvardsen T, Torp H, Crosby J, Lyseggen E,et al. Noninvasive myocardial strain measurement by speckle tracking echocardiography: validation against sonomicrometry and tagged magnetic resonance imaging. J Am Coll Cardiol 2006;47:789-93″
Tagging seems to be explored a lot in NYU, Penn State University, and also Johns Hopkins University (there is group outside the United States which works on this area as well, but further reading will probably reveal many more such groups). N.F. Osman and J.L. Prince from Johns Hopkins University, for example, wrote about “Angle images for measuring heart motion from tagged MRI” (presented in the 1998 International Conference on Image Processing in Chicago, Illinois). The abstract says that their work “introduces a new image processing technique for rapid analysis and visualization of tagged cardiac magnetic resonance (MR) images. The method is based on the use of isolated spectral peaks in SPAMM-tagged magnetic resonance images. The authors call the phase of an image corresponding to just one of these peaks an angle image, and show that except for a phase-wrapping artifact, an angle image is linearly related to a component of the three-dimensional motion. Using one or more angle images, the authors show how to synthesize conventional tag lines, reconstruct displacement fields for small motions, calculate the optical flow between successive temporal images, and calculate two-dimensional strain. The authors demonstrate the performance of this approach on both real and simulated tagged MR images”
Ten years later, 4-D work (including temporal) was done by the group from NYU School of Medicine along with Dimitris N Metaxas and Zhen Qian [2] (from the Center for Computational Biomedicine Imaging and Modeling at Rutgers University). They published “A Segmentation and Tracking System for 4D Cardiac Tagged MR Images” (issue date of 2006, with current version from 2008). Their paper from EMBS ’06 (28th Annual International Conference of the IEEE) “present[s] a robust method for segmenting and tracking cardiac contours and tags in 4D cardiac MRI tagged images via spatio-temporal propagation. Our method is based on two main techniques: the Metamorphs segmentation for robust boundary estimation, and the tunable Gabor filter bank for tagging lines enhancement, removal and myocardium tracking.”
The Institute of Biomedical Engineeering at Bogazici University (Istanbul, Turkey) has also published [3] “Towards rapid screening of tagged MR images of the heart” and the abstract reads: “The final aim of this work is to perform rapid classification of tagged cardiac MR images as normal and abnormal. In the proposed technique, images are first analyzed using harmonic phase analysis and synthetic tags are computed over the myocardium. Cubic curves are fitted to these tags and curve parameters are compared at various regions of the myocardium. In this initial study, the ratios of curve parameters between normal and diseased hearts, such as dilated cardiomyopathy (DCM) and heart with infarcted regions, are evaluated. If the initial segmentation problems are solved, this method could be a very fast and automatic screening tool for identifying diseased locations in tagged MRI.”
Background to My Work
A while ago I began implementing an approach where I explore new methods of anatomical structures extraction based on tagging. After spending a long time trying to get my hands on some data with tagging in it (low-resolution videos is the best I have found on the Web so far [1, 2, 3]) I managed to find MRI cardiac data which the people from New York can share with us. Getting data with tagging has proven tricky unless there is direct access to the right people and I’m grateful to receive some good data as I’m eager to work on extending and complementing the work which is Axel’s breakthrough. Doing so without the required data would be hard. One possibility for working with such data is to locally work on sythesising or emulating a set of tagged image using specific software which does this quite nicely. It may be good if we need to test programs where the ground truth solution is known. Otherwise, approaching the group from New York was a reasonable option to be exploring and with enough data, both real and synthetic data can be used, one for actual work and another for validation of the method.
Why Shuffle Distance/Difference
In a later short paper I’ll explain the approach of encoding the image in terms of bytes (8 bits) or blocks of 64 bits that can be handled by particular CPUs more efficiently and thus have an image stored and processed (even in 3-D) with little burden on the hardware, yielding faster results for real-time tracking of the tags. There is an existing implementation left from my work on brain images where this is done in a vectorised fashion and works quite fast in Octave (bar JIT) or MATLAB. Whether tagging in 3-D is available at all remains to be seen. That is a subject for another day.
References:
[1] N.F. Osman, J.L. Prince, “Angle images for measuring heart motion from tagged MRI,” International Conference on Image Processing, vol. 1, pp.704, 1998.
[2] D. N. Metaxas, L. Axel, Z. Qian, and X. Huang, “A Segmentation and Tracking System for 4D Cardiac Tagged MR Images,” Engineering in Medicine and Biology Society, pp. 1541–1544, 2006.
[3] D. Goksel, M. Ozkan, and C. Ozturk, “Towards rapid screening of tagged MR images of the heart,” Engineering in Medicine and Biology Society, vol. 1, pp. 156–159, 2001.






 Filed under:
Filed under: