Keywords: Open Access; Free software; Cardiac cine-MRI; Brain; Segmentation; Tagging; Cardiac deformation analysis
Overview
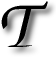 his informal post presents a quick survey of what’s available for one to find when it comes to heart tagging downloads (without having to go through radiologists and all sorts of burdensome forms). It is a problem I’ve been tackling recently and I would like to share my findings to make life easier for other scientists who find themselves in the same situation.
his informal post presents a quick survey of what’s available for one to find when it comes to heart tagging downloads (without having to go through radiologists and all sorts of burdensome forms). It is a problem I’ve been tackling recently and I would like to share my findings to make life easier for other scientists who find themselves in the same situation.
Some Background Information
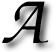 WEEK OR SO ago I decided to share information about the availability of datasets for researchers. When faced with the problem of looking for, say, ‘”tagged MR” images heart download dicom’ (and a lot of searching around this phrase) I was surprised to find that almost nobody publicly shared or made discoverable a dataset one can use for research purposes. In order to simplify the task for others who find themselves in a similar situation, several days ago I wrote about my intentions to make a sort of summary [1, 2].
WEEK OR SO ago I decided to share information about the availability of datasets for researchers. When faced with the problem of looking for, say, ‘”tagged MR” images heart download dicom’ (and a lot of searching around this phrase) I was surprised to find that almost nobody publicly shared or made discoverable a dataset one can use for research purposes. In order to simplify the task for others who find themselves in a similar situation, several days ago I wrote about my intentions to make a sort of summary [1, 2].
As a bit of background, our original plan was to write and implement special protocols with engineers at the MRI scanner, as part of the research effort by which special types of data are obtained for a novel approach to be tested. This is explained in a paper I’ll put up in the site pretty soon.
At the moment, due to particular constraints, we are working on analysis of sequences of tagged data. This obviously requires not just MR images of the heart but also sequences and these, preferably sequences that must have particular types of tags which a radiologist would use. Data can be acquired/sliced from different angles and obtained from any subject/patient at the hospital*, but for this particular approach that we test (to be covered in a separate post) this is rather irrelevant as we want it to be applicable regardless of factors like angles, subjects, and potentially their atrophies.
Resources
SourceForge hosts quite a few MRI-related software projects which are free/libre and thus of value to MRI-based research (our methods and data should be made freely accessible for independent validation). One of these projects is the cardiac MR toolbox for Matlab, which may also be fully compatible with Octave (free/libre software). “The cardiac MR (CMR) toolbox for Matlab,” explains the official Web page, “provides tools for image registration, perfusion analysis, segmentation, and image reconstruction from sparsely sampled data.” Here is the alternative project homepage. It states:
This is the Cardiac MR toolbox for Matlab project (“cmr-toolbox”)
This project was registered on SourceForge.net on Mar 29, 2010, and is described by the project team as follows:
The cardiac MR (CMR) toolbox for Matlab provides tools for image registration, perfusion analysis, segmentation, and image reconstruction from sparsely sampled data.”
For brain MRI there is the RESCUE brain MRI segmentation project. It is not yet available, but it soon will be. Its author states: “RESCUE will be made available after publication of the PhD thesis of James Withers (Uni of Edinburgh, UK). The first version of the package will initially include super-resolution, thin structure detection, and partial volume estimation components.”
Of course, one can get brain datasets from the rather famous BrainWeb (more here) and some more data resides in my site, including a sample set of images and some DICOM-formatted data of my own brain (acquired at Hope Hospital for a colleague’s experiment back in 2005).
Moving on to the heart, there is a cardiac MRI dataset which is not terribly useful because almost everything is encoded and stored in MATLAB format and may therefore require some loaders that are proprietary (Octave does not cope so well with those).
There is another project called inTag and this Web pageoffers a a look at what’s done there. Another page offers a look at how it’s used. It is “a software to calculate, display and analyze myocardial strains and intra-myocardial mechanics from cardiac MR images with a tagging pattern.”
The dependencies on proprietary software are unfortunate though. As the page puts it: “After a preliminary prototyping step in Matlab ®, InTag is now available as a plugin in OsiriX, the advanced open-source image processing software. It benefits of all its unique capabilities for navigation, visualisation of multimodality and multidimensional images, and data management.”
This requires OsiriX, which is Free/open source software, but only for Macs, which is not terribly useful then. Here is some tagging from this software package called inTag. It’s just a group of some very low-resolution images in DICOM format (small and not so useful at all).
Another dataset which I find odd (not because it’s made available just in very low resolution but because there is something odd in orientation) comes from New Zealand. Images are said to be available for download, but they are all encoded as AVI-formatted slanted videos and also compressed. The only one set with tagging data is not too helpful, either. The page states that “The Auckland MRI Research group has agreed to make freely available 256×256 JPEG images and image orientation information for download. In addition the Auckland MRI Research group has also agreed to make freely available the cines in AVI format”. What’s listed in the page as an overview is as follows:
All 119 T1-weighted jpeg images
akld-mrirg-atlas-jpegs.zip (4.33MB)
12 Long axis trueFisp avi movies
akld-mrirg-cines-truefisp-long.zip (21.76MB)
1-15 Short axis trueFisp avi movies
akld-mrirg-cines-truefisp-short-a.zip (25.32MB)
16-30 Short axis trueFisp avi movies
akld-mrirg-cines-truefisp-short-b.zip (28.61MB)
3 Aorta and 1 RVOT trueFisp avi movies
akld-mrirg-cines-truefisp-aorta-rvot.zip (7.42MB)
11 Long axis and 15 Short axis tagged avi movies
akld-mrirg-cines-tagging.zip (26.46MB)
Aorta(2) / Right(2) and Left(2) Pulmonary arteries(4) phase contrast avi
movies akld-mrirg-cines-phasecontrast.zip (10.34MB)
Those sets are not tagged except akld-mrirg-cines-tagging.zip. There is also no data from the Cincinnati Children’s Hospital Medical Center, which shows up among many search results. Search for data has proven to be quite fruitless, generally speaking.
Eventually I used data which I acquired from someone’s thesis frame by frame, only to be seen as a temporary solution until data comes from a group that keeps it for internal use. The conclusion is that the Web is generally a poor resource for such data, especially if tagging is a requirement. If someone knows of an exception, please comment below.
Some References
On this fruitless pursuit for data the following references were seen as potentially relevant (publication names mostly omitted as it’s easy to search by title).
DN Metaxas, “A Segmentation and Tracking System for 4D Cardiac Tagged MR Images,” 2008.
L Axel, “Tagged MRI-Based Studies of Cardiac Function,” Computerized Medical Imaging and Graphics, Volume 29, Issue 8, December 2005, Pages 607-616, 2003.
D Goksel, “Towards rapid screening of tagged MR images of the heart,” 2002.
NF Osman, “Angle Images for Measuring Heart Motion from Tagged MRI.”
L Axel, “An Integrated Program for 2-D and 3-D Analysis of Heart Wall…,” 2002.
Y. Chenounea, “Segmentation of cardiac cine-MR images and myocardial deformation assessment using level set methods,” 2005.
(Abstract: “In this paper, we present an original method to assess the deformations of the left ventricular myocardium on cardiac cine-MRI. First, a segmentation process, based on a level set method is directly applied on a 2D+t dataset to detect endocardial contours. Second, the successive segmented contours are matched using a procedure of global alignment, followed by a morphing process based on a level set approach. Finally, local measurements of myocardial deformations are derived from the previously determined matched contours. The validation step is realized by comparing our results to the measurements achieved on the same patients by an expert using the semi-automated HARP reference method on tagged MR images.)
____
* Some are too blurry and others paint areas with blood presence black, which help isolate the boundaries of important structures for subsequent analysis.

 Y EARLIER experiences with Konqueror go back to ~2002 when I tried it at work under Mandrake, around the early versions of KDE3 or later KDE2. Konqueror had many menu options and was daunting in some ways compared to Netscape and Mozilla. In KDE 3.1 Konqueror had acceptance problems among Web sites that were simply IE-centric or IE- and Netscape-centric. This was not a problem in Konqueror itself, but it made life a little harder for Konqueror users like myself. I only moved to Firefox some time in 2004 and it is still my Web browser of choice, having tested Chromium and some other browsers for a while (they lack plugins).
Y EARLIER experiences with Konqueror go back to ~2002 when I tried it at work under Mandrake, around the early versions of KDE3 or later KDE2. Konqueror had many menu options and was daunting in some ways compared to Netscape and Mozilla. In KDE 3.1 Konqueror had acceptance problems among Web sites that were simply IE-centric or IE- and Netscape-centric. This was not a problem in Konqueror itself, but it made life a little harder for Konqueror users like myself. I only moved to Firefox some time in 2004 and it is still my Web browser of choice, having tested Chromium and some other browsers for a while (they lack plugins).





 Filed under:
Filed under: 
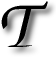 AGGING in the context of image acquisition involves getting magnetic signal by selectively sending charges to the tissue, whose atoms (usually just hydrogen is targeted by adjustment of frequencies) equipment can try to test for response in such a way that under normal conditions without noticeable change they would return a rectangular grid overlaid on top of the data. This enables better tracking of tissue motion that’s robust to spatially-similar atoms (whose returned signal is hard to discern visually). This post is a personal and informal quick survey of some resources which people may hopefully find helpful. It is not a detailed or comprehensive analysis.
AGGING in the context of image acquisition involves getting magnetic signal by selectively sending charges to the tissue, whose atoms (usually just hydrogen is targeted by adjustment of frequencies) equipment can try to test for response in such a way that under normal conditions without noticeable change they would return a rectangular grid overlaid on top of the data. This enables better tracking of tissue motion that’s robust to spatially-similar atoms (whose returned signal is hard to discern visually). This post is a personal and informal quick survey of some resources which people may hopefully find helpful. It is not a detailed or comprehensive analysis.

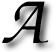 WEEK OR SO ago I decided to share information about the availability of datasets for researchers. When faced with the problem of looking for, say, ‘”tagged MR” images heart download dicom’ (and a lot of searching around this phrase) I was surprised to find that almost nobody publicly shared or made discoverable a dataset one can use for research purposes. In order to simplify the task for others who find themselves in a similar situation, several days ago I wrote about my intentions to make a sort of summary [
WEEK OR SO ago I decided to share information about the availability of datasets for researchers. When faced with the problem of looking for, say, ‘”tagged MR” images heart download dicom’ (and a lot of searching around this phrase) I was surprised to find that almost nobody publicly shared or made discoverable a dataset one can use for research purposes. In order to simplify the task for others who find themselves in a similar situation, several days ago I wrote about my intentions to make a sort of summary [